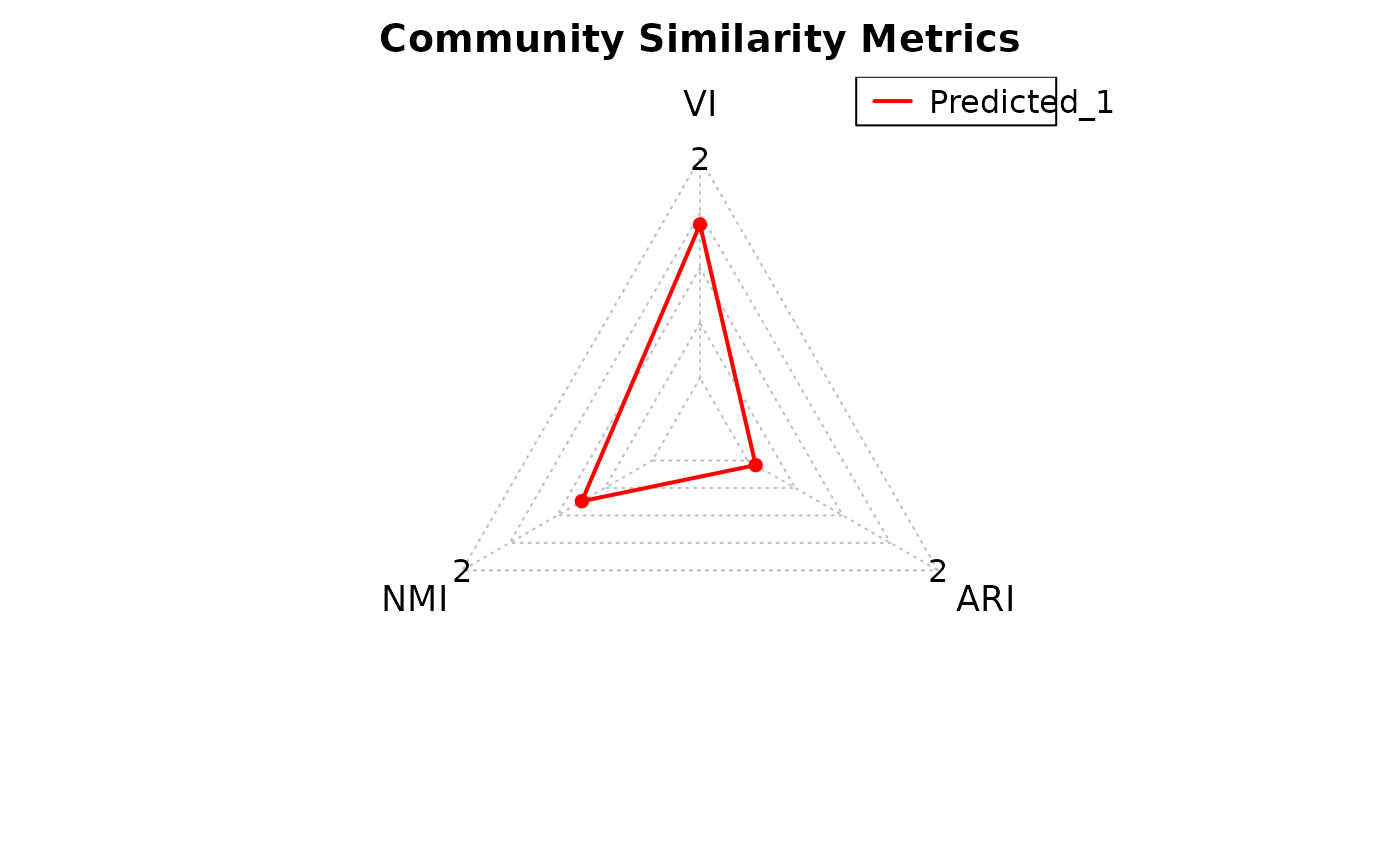
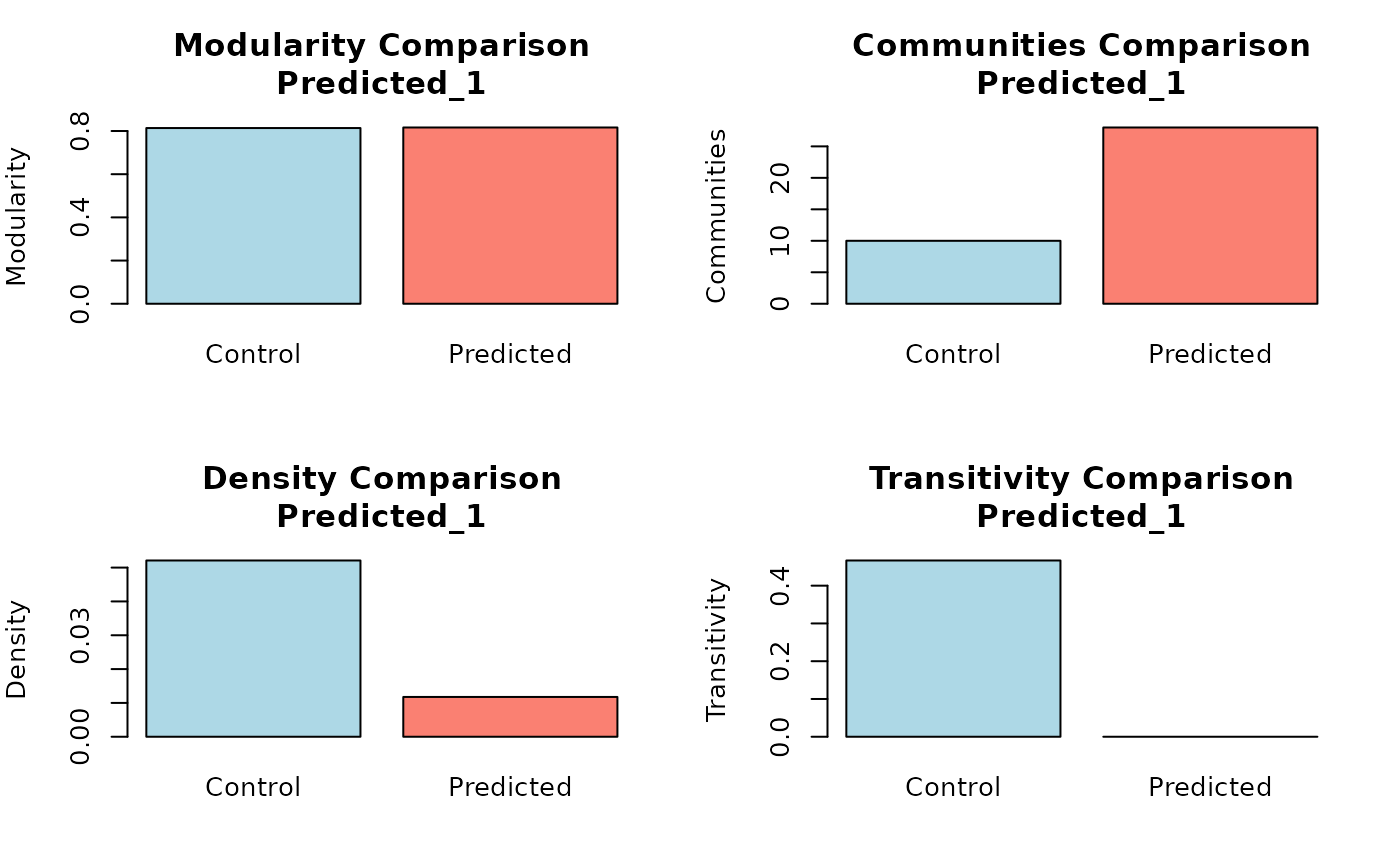
Compare Community Assignments and Topological Properties
Source:R/community_similarity.R
community_similarity.RdConvenience wrapper that computes community assignment metrics,
topological properties, and optionally visualizes the comparison.
For more control, use the individual functions:
compute_community_metrics,
compute_topology_metrics, and
plot_community_comparison.
Arguments
- control_output
A list output from
community_path()representing the ground truth network. Must contain agraph(igraph object) andcommunities$membership.- predicted_list
A list of lists, each output from
community_path()representing predicted networks to compare.- plot
Logical. If TRUE, displays plots immediately. If FALSE, no plots are displayed. Default: TRUE.
Value
A list containing:
community_metrics: A data frame with VI, NMI, and ARI scores for each prediction.topology_measures: A data frame with raw topological metrics for each prediction.control_topology: A list of raw topological metrics for the ground truth network.
Details
This function requires the igraph package. If
plot = TRUE, the fmsb package is also required.
Community similarity is measured using variation of information (VI),
normalized mutual information (NMI), and adjusted Rand index (ARI).
Examples
data(toy_counts)
data(toy_adj_matrix)
# Infer networks (toy_counts is already a MultiAssayExperiment)
networks <- infer_networks(
count_matrices_list = toy_counts,
method = "GENIE3",
nCores = 1
)
head(networks[[1]])
#> regulatoryGene targetGene weight
#> 1 HLA-B FTL 0.2069098
#> 2 HLA-A HLA-B 0.1617015
#> 3 CD74 CXCR4 0.1564308
#> 4 HLA-B HLA-A 0.1498623
#> 5 FTL FTH1 0.1445262
#> 6 FTH1 FTL 0.1410349
# Generate adjacency matrices
wadj_se <- generate_adjacency(networks)
swadj_se <- symmetrize(wadj_se, weight_function = "mean")
# Apply cutoff
binary_se <- cutoff_adjacency(
count_matrices = toy_counts,
weighted_adjm_list = swadj_se,
n = 1,
method = "GENIE3",
quantile_threshold = 0.95,
nCores = 1,
debug = TRUE
)
#> [Method: GENIE3] Matrix 1 → Cutoff = 0.06771
#> [Method: GENIE3] Matrix 2 → Cutoff = 0.06327
#> [Method: GENIE3] Matrix 3 → Cutoff = 0.06936
head(binary_se[[1]])
#> [1] "ACTG1" "ARPC2" "ARPC3" "BTF3" "CD3D" "CD3E"
consensus <- create_consensus(binary_se, method = "union")
comm_cons <- community_path(consensus)
#> Detecting communities...
 #> Running pathway enrichment...
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
comm_truth <- community_path(toy_adj_matrix)
#> Detecting communities...
#> Running pathway enrichment...
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
comm_truth <- community_path(toy_adj_matrix)
#> Detecting communities...
 #> Running pathway enrichment...
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
sim_score <- community_similarity(comm_truth, list(comm_cons))
#> Running pathway enrichment...
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
sim_score <- community_similarity(comm_truth, list(comm_cons))